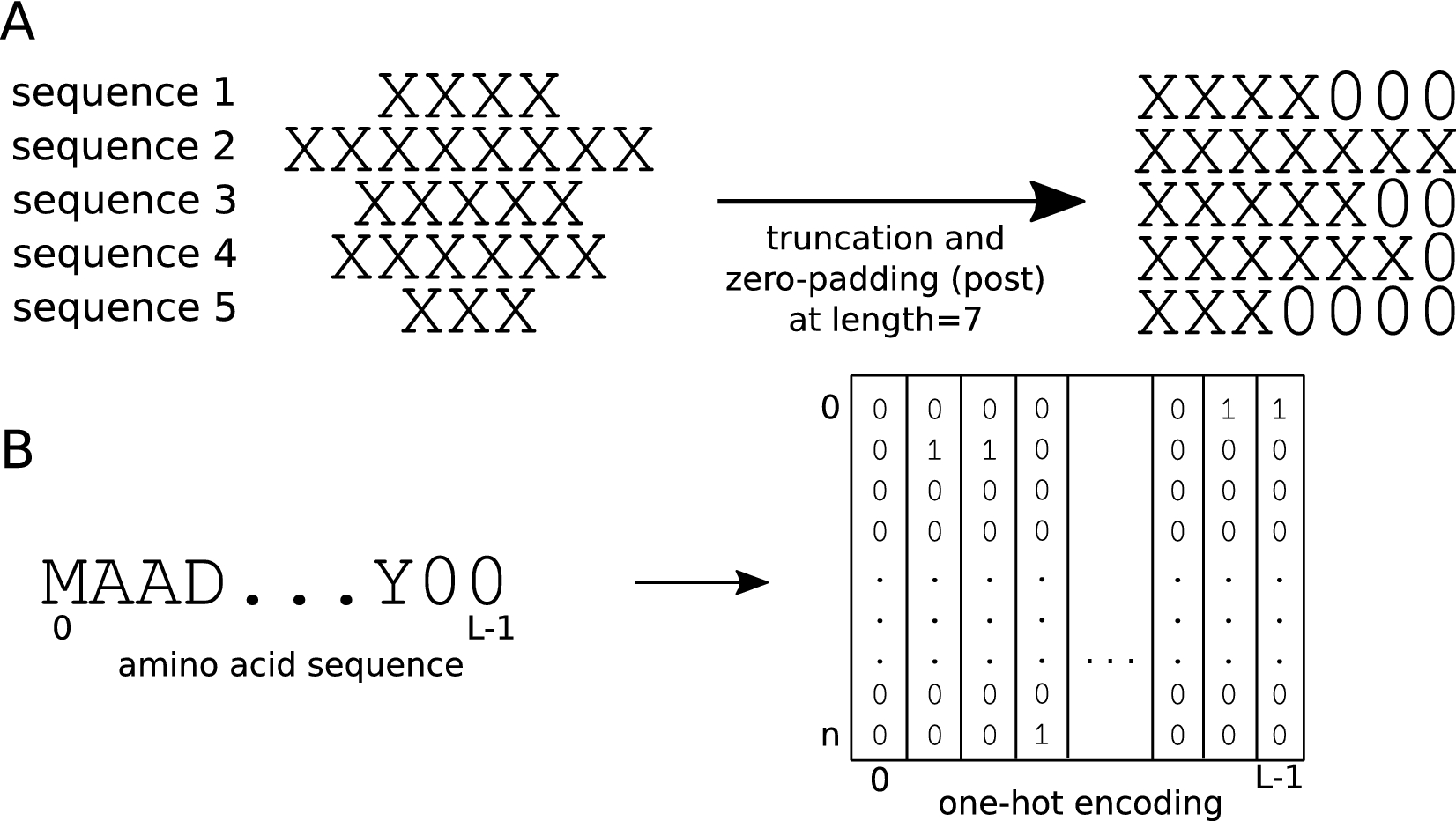
Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction
By A Mystery Man Writer
Sensors, Free Full-Text

Nucleotide-level Convolutional Neural Networks for Pre-miRNA Classification

Amino Acid Encoding Methods for Protein Sequences: A Comprehensive Review and Assessment

MECE: a method for enhancing the catalytic efficiency of glycoside hydrolase based on deep neural networks and molecular evolution - ScienceDirect

A novel technique for multiple failure modes classification based on deep forest algorithm

Dokumen - Pub Machine Learning in Bioinformatics of Protein Sequences Algorithms Databases and Resources For Modern Protein Bioinformatics 9811258570 9789811258572, PDF, Protein Structure

Using Deep Learning to Annotate the Protein Universe

MECE: a method for enhancing the catalytic efficiency of glycoside hydrolase based on deep neural networks and molecular evolution - ScienceDirect
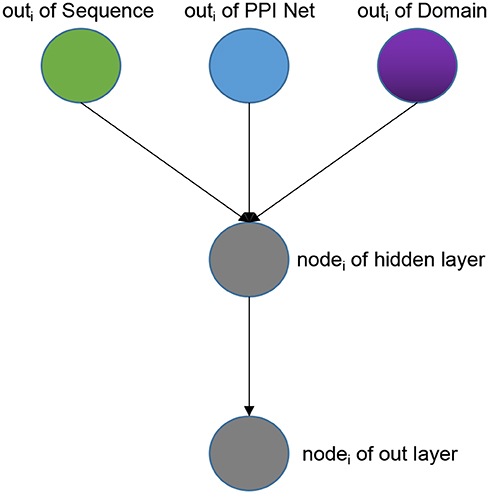
Frontiers SDN2GO: An Integrated Deep Learning Model for Protein Function Prediction

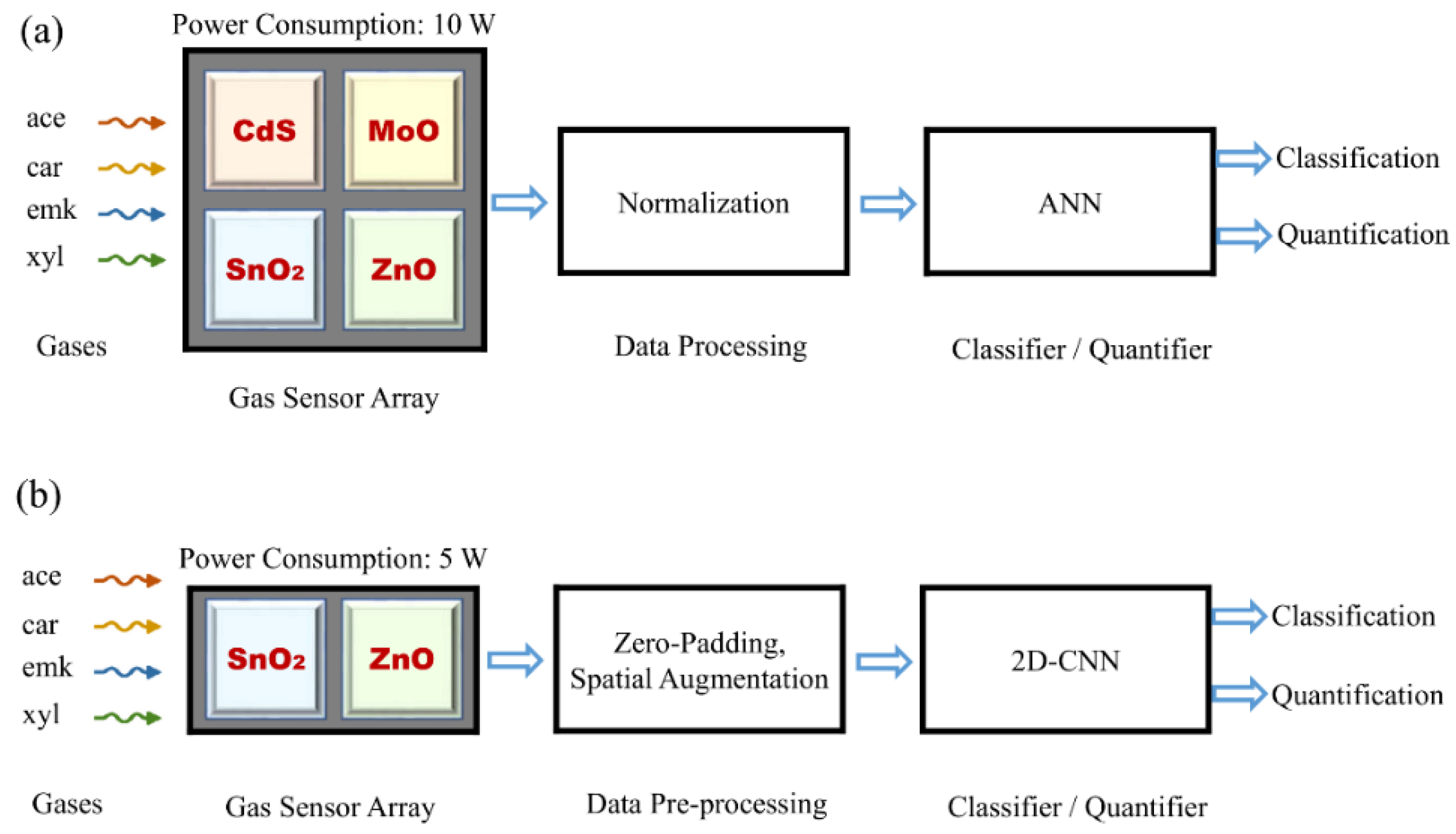
PDF) Zero-Padding and Spatial Augmentation-Based Gas Sensor Node Optimization Approach in Resource-Constrained 6G-IoT Paradigm

Battery degradation prediction against uncertain future conditions with recurrent neural network enabled deep learning - ScienceDirect

Effect of sequence padding on the performance of deep learning models in archaeal protein functional prediction

Codon language embeddings provide strong signals for use in protein engineering
- Fixed navigations and sections - here is scroll-padding - DEV Community

- Adjust image padding in an email - Constant Contact Community - 391629
- Padding Gasket for Flanges and Clamps - Filcoflex

- Flexible Wholesale Jacket Padding Materials For Clothing And More

- Fjallraven Skogso Padded Jacket (Black) – WeyFarm Outdoors

- Closeup view of slim woman in underwear on white background. Cellulite problem concept Stock Photo

- Falke Shelina 12 denier Pantyhose Stockings, Women's Fashion

- Non-Padded Gol Chakri Back X Sports Bra at Rs 62/piece in

- Bryson Born Shoes

- Buy StyFun Women Multicolor Solid Cotton Blend Pack Of 2 Bra & Panty Set Online at Best Prices in India - JioMart.
)
