GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
By A Mystery Man Writer
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
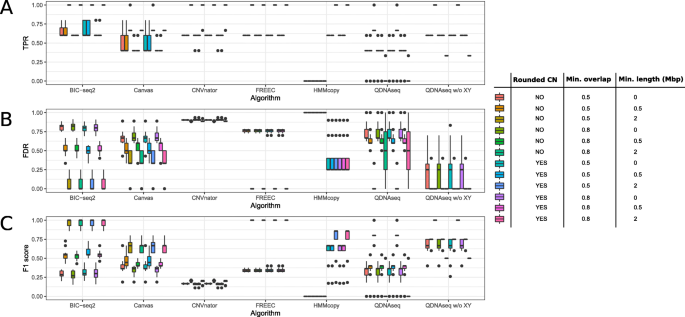
Evaluation of tools for identifying large copy number variations from ultra- low-coverage whole-genome sequencing data, BMC Genomics
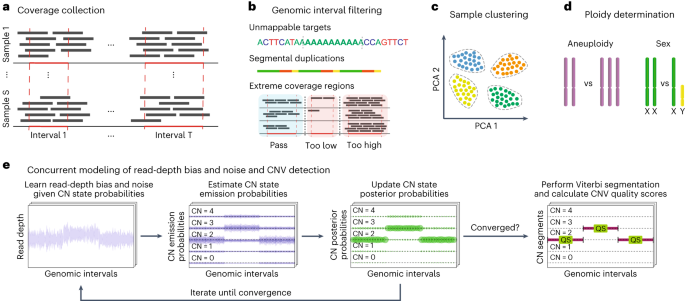
GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data

Shallow whole-genome sequencing of plasma cell-free DNA accurately differentiates small from non-small cell lung carcinoma, Genome Medicine
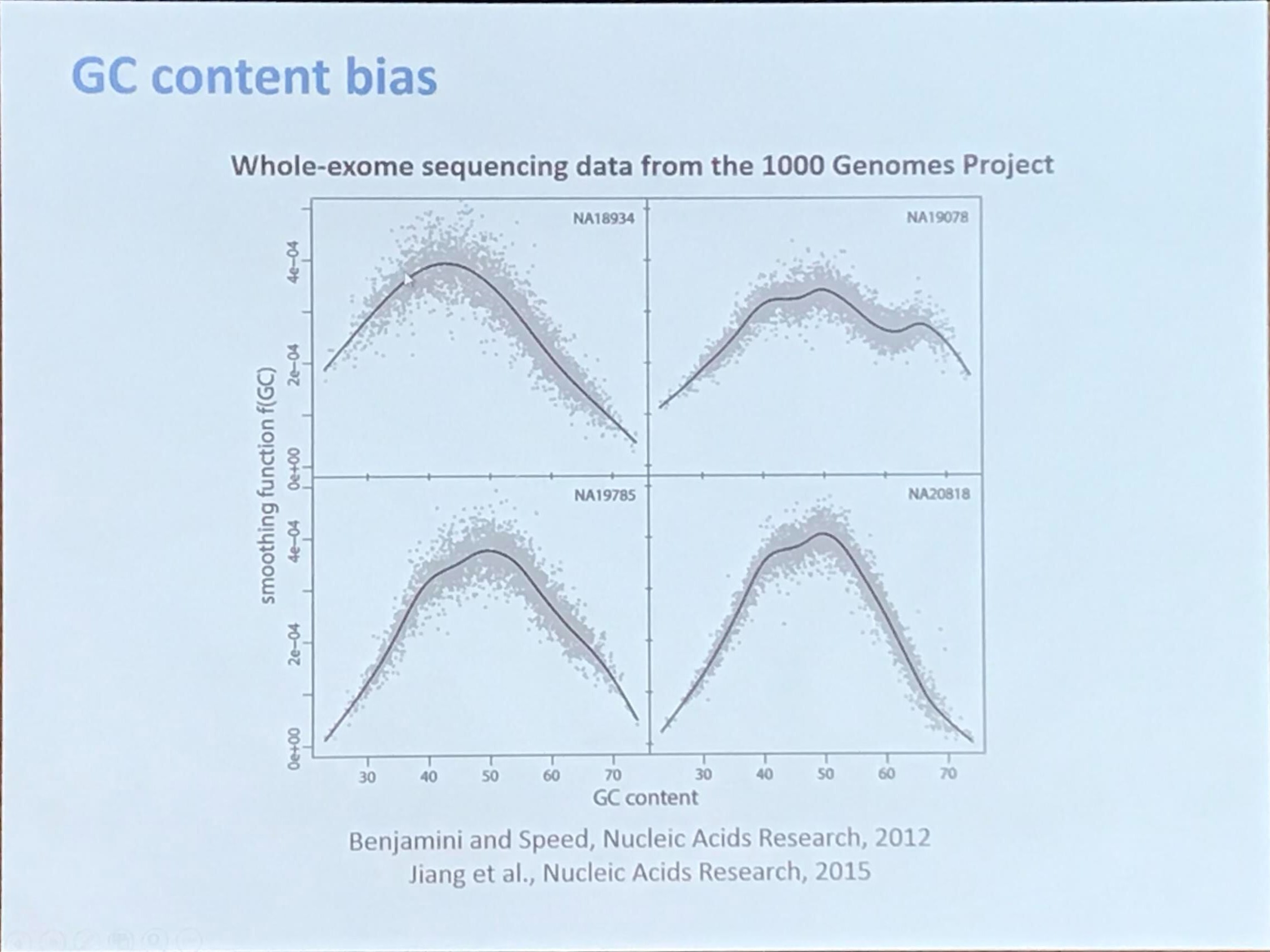
DNA copy number profiling: from bulk tissue to single cells

CODEX2: full-spectrum copy number variation detection by high-throughput DNA sequencing, Genome Biology

Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach

PDF) Low-Coverage Whole Genome Sequencing Using Laser Capture Microscopy with Combined Digital Droplet PCR: An Effective Tool to Study Copy Number and Kras Mutations in Early Lung Adenocarcinoma Development
Tumor Genome Analysis Core · GitHub
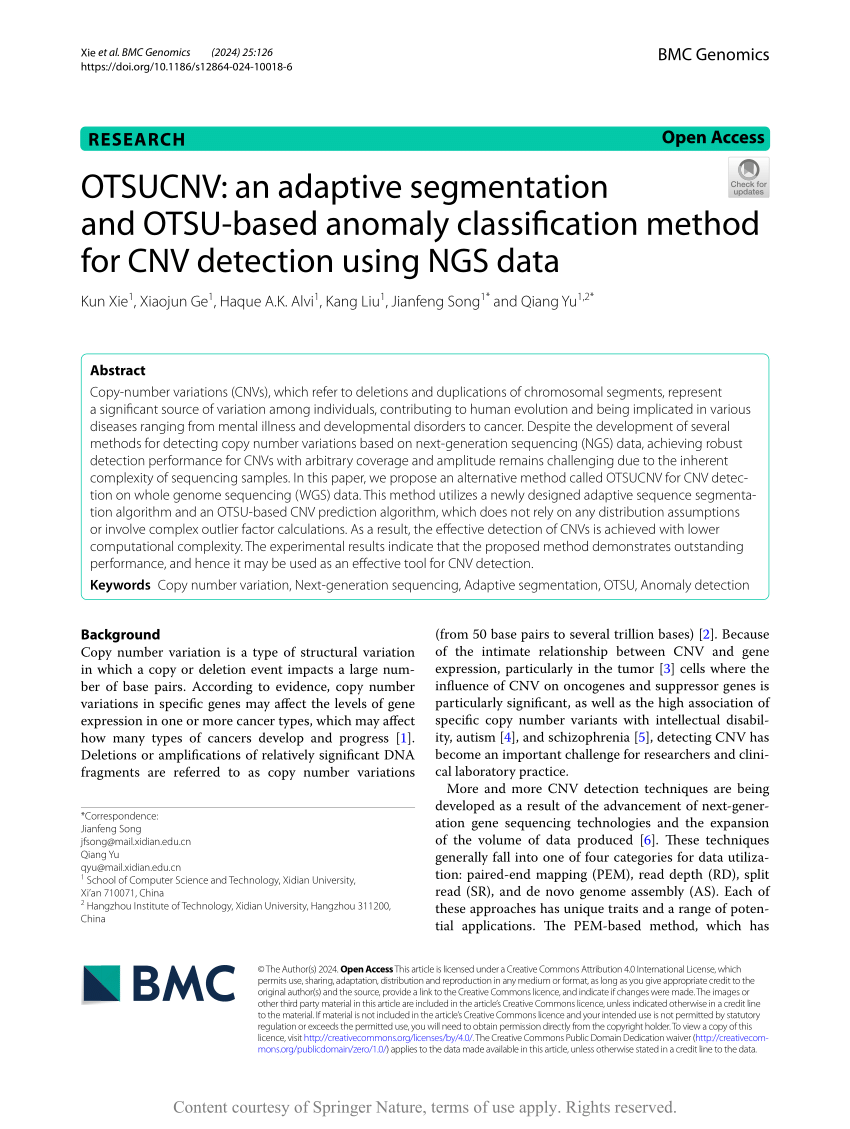
PDF) OTSUCNV: an adaptive segmentation and OTSU-based anomaly classification method for CNV detection using NGS data
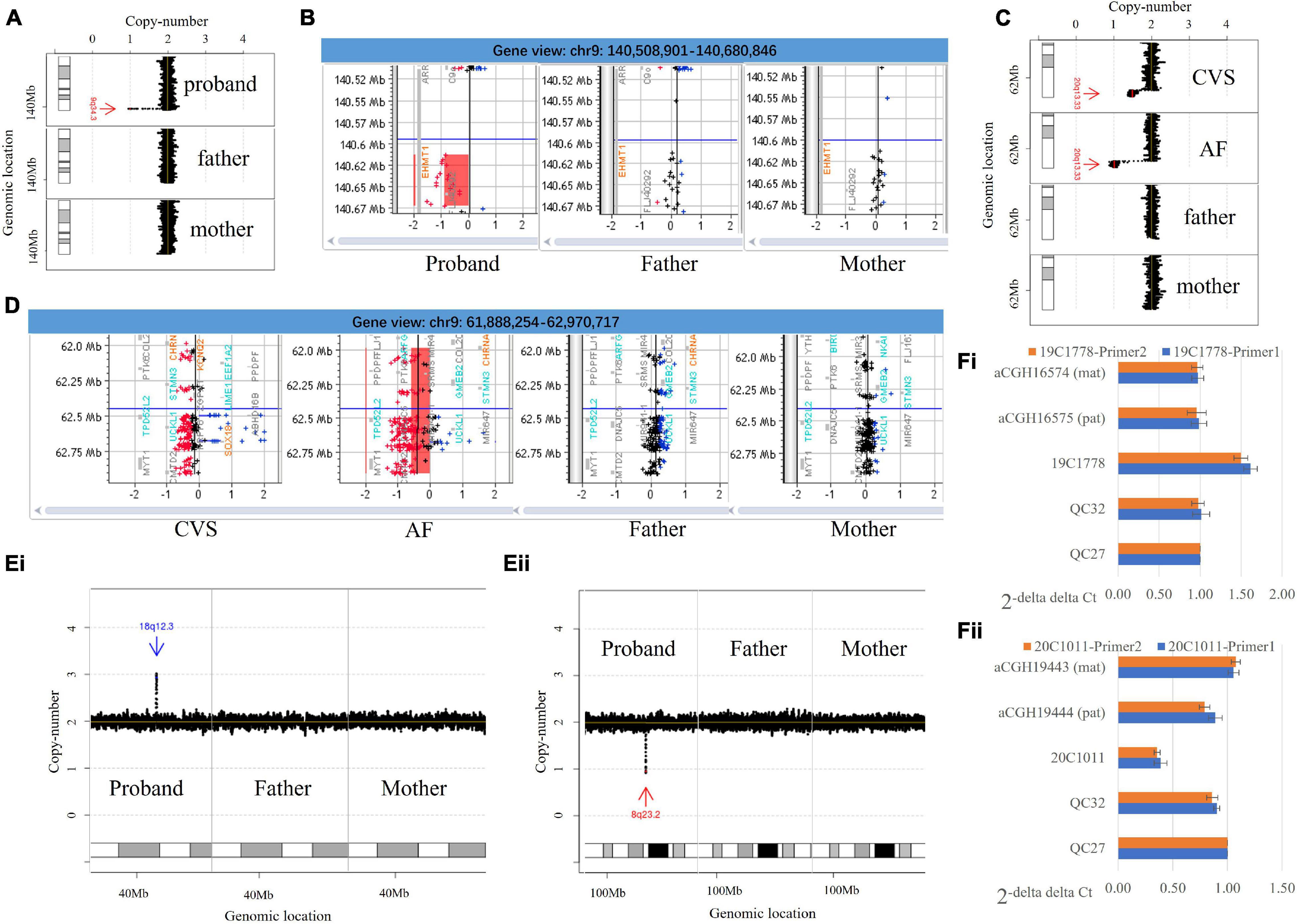
Frontiers Trio-Based Low-Pass Genome Sequencing Reveals Characteristics and Significance of Rare Copy Number Variants in Prenatal Diagnosis

Copy number analysis by low coverage whole genome sequencing using ultra low-input DNA from formalin-fixed paraffin embedded tumor tissue, Genome Medicine

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data

Copy number profiles are stable. Ultra-low pass whole genome sequencing
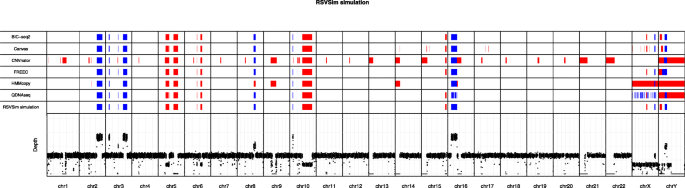
Evaluation of tools for identifying large copy number variations from ultra- low-coverage whole-genome sequencing data, BMC Genomics

PDF) CNHplus: the chromosomal copy number heterogeneity which respects biological constraints
- Low Coverage

- Low-coverage whole-genome sequencing of cerebrospinal-fluid-derived cell-free DNA in brain tumor patients - ScienceDirect

- GitHub - Rosemeis/emu: EM-PCA for Ultra-low Coverage Sequencing Data
- Low-E glass affect 5G/4G wireless coverage

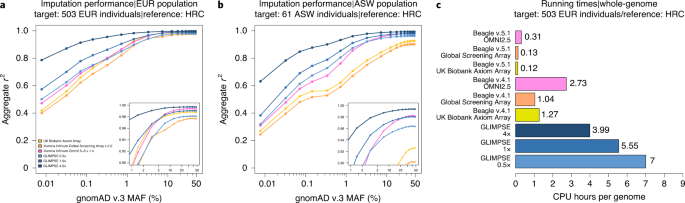
- Efficient phasing and imputation of low-coverage sequencing data
- XFLWAM Women’s Casual Baggy Sweatpants High Waisted Running Joggers Pants Athletic Trousers with Pockets Drawstring Track Pants Black S

- 6732 MAGIC CURVES SEAMLESS FULL BODY LONG LEG (6pcs Wholesale

- Iron Bull Strength Powerlifting Belt - 10mm Double Prong - 4-inch Wide - Heavy Duty for Extreme Weight Lifting Belt (All Black, Small) : Sports & Outdoors

- FOREVER MAXI TANK DRESS- ATLANTIS

- Angel Graphic Cropped Tank Top

- Amoena Nina Underwire Bra, Size 42d, Nude #56789056

- 5 Tips for Choosing the Correct Crochet Hook Size, Crochet

- The Plumber's Choice 1 in. Brass Press Y-Strainer Valve 322S327-NL

- 6 Kilograms Huge Hand Knit Wool Sweater Made of 100 % Soft Wool T1094 - Sweden

- Hormonal IUDs and What They Mean For Your Endometrial Lining